Applications
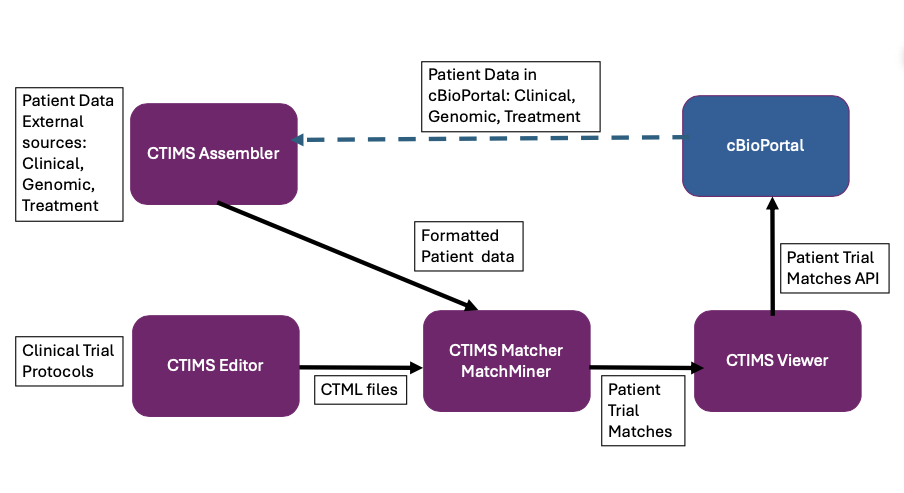
CTIMS consists of the following components.
CTIMS has four components namely Assembler, Editor, Matcher and Viewer. The Assembler converts study data from cBioPortal format to MatchMiner format, either from locally saved cBioPortal study files or by retrieving data via the cBioPortal API. CTIMS Editor helps to create CTMLs efficiently and store them in a database. CTIMS Matcher is the DFCI MatchMiner Engine. DFCI MatchMiner uses CTML files to find patient trial matches. CTIMS Editor integrates with MatchMiner via MatchMiner API and Message Queue for supporting QuickMatch. QuickMatch allows the curator to quickly verify the CTML. CTIMS Viewer stores the match results for data analysis and visualization. CTIMS Viewer API supports integration with cBioPortal so that the trial match results can be viewed in cBioPortal along with patient data. CTIMS supports Authentication and Authorization using Keycloak.